|
The Transposons We Study
Our
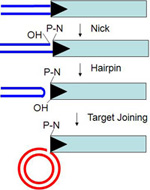
research is focused on several different DNA cut-and-paste transposons that move
by excision from the donor site by double-strand breaks at each end of the
element, followed by joining of these newly exposed transposon ends to a new
insertion site. Most such transposable elements display limited site
selectivity, inserting into many different target sites. One element that we
study, however, the bacterial transposon, Tn7, displays unusual target
selectivity.
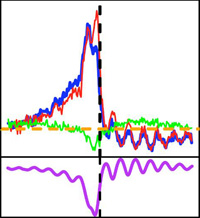 The other elements we study are members of the widespread eukaryotic hAT and
piggyBac superfamilies; there are active members of these families in
vertebrates. We were originally attracted to the study of these elements because
nothing was known about their transposition at the molecular level. Many
genomes, including the human genome, also contain intact hAT and piggyBac
transposase-like proteins whose functions interest us. The other elements we study are members of the widespread eukaryotic hAT and
piggyBac superfamilies; there are active members of these families in
vertebrates. We were originally attracted to the study of these elements because
nothing was known about their transposition at the molecular level. Many
genomes, including the human genome, also contain intact hAT and piggyBac
transposase-like proteins whose functions interest us.
We study several hAT elements: Hermes, an element closely related to the hobo
element, which has been studied in Drosophila; AeBuster1, the first active
transposon to be found in the mosquito Aedes aegypti, the vector of malaria;
TcBuster, an element from the red beetle Tribolium castaneum that we have found
is highly active in mammalian cells; Tol2, a medaka fish element that can also
transpose in zebrafish and mammalian cells; and SPIN, a now inactive family of
elements that appears to have spread though some mammalian genome by horizontal
transfer that we have “resurrected” by making a consensus derivative.
We also study several piggyBac elements: the Trichoplusia ni piggyBac from the
cabbage looper moth, which has been used for transgenesis in many other
organisms, and Myotis lucifugus piggyBac, an element from the little brown bat
that we identified and is the first active DNA transposon found in a mammalian
genome. Previous studies found no evidence that DNA cut-and-paste elements have
been active in the past 35 million years in mammalian genomes.
Our Research Strategies
 To
probe the transposition mechanisms of the elements we study, we have developed
in vitro transposition systems reconstituted with purified proteins. We have
found that common chemical mechanisms underlie the movement of all of these
elements and that the active-site regions of their transposases are actually
intimately related in structure. The similarities between these elements reflect
the fact that the breakage and joining reactions that underlie the movement of
all DNA cut-and-paste elements and the DNA forms of retroviruses are highly
similar and that their transposases and integrases are structurally related.
These in vitro systems allow us to further dissect transposition at the
molecular level. We have also established heterologous transposition systems for
most of our hAT and piggyBac elements in the budding yeast Saccharomyces
cerevisae. The genetic tractability of yeast has allowed us to isolate mutant
versions of our transposons, such as hyperactive versions, by in vivo screening
and also to ask how the host influences transposition. We are exploiting the
yeast gene knockout collections in these studies of host factors. To
probe the transposition mechanisms of the elements we study, we have developed
in vitro transposition systems reconstituted with purified proteins. We have
found that common chemical mechanisms underlie the movement of all of these
elements and that the active-site regions of their transposases are actually
intimately related in structure. The similarities between these elements reflect
the fact that the breakage and joining reactions that underlie the movement of
all DNA cut-and-paste elements and the DNA forms of retroviruses are highly
similar and that their transposases and integrases are structurally related.
These in vitro systems allow us to further dissect transposition at the
molecular level. We have also established heterologous transposition systems for
most of our hAT and piggyBac elements in the budding yeast Saccharomyces
cerevisae. The genetic tractability of yeast has allowed us to isolate mutant
versions of our transposons, such as hyperactive versions, by in vivo screening
and also to ask how the host influences transposition. We are exploiting the
yeast gene knockout collections in these studies of host factors.
Hyperactive mutants are of interest because they can reveal how transposition is
controlled. They also provide better tools for genome engineering, such as
insertional mutagenesis and transgenesis. We have isolated both hAT and piggyBac
mutants in yeast that are also hyperactive in mammalian cells, including stem
cells.
 In contrast to the single transposition protein encoded by hAT and piggyBac
elements, four Tn7-encoded proteins mediate the high-frequency insertion of Tn7
into a specific chromosomal site called attTn7. Tn7 transposition requires the
assembly of an elaborate nucleoprotein complex containing both the donor DNA
from which Tn7 excises and the target DNA into which Tn7 inserts, which contains
all the Tns proteins. Our goal is to understand the structure of this complex
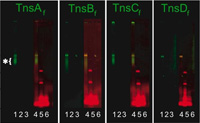
and how its activity is regulated. We now use in vitro assays to demonstrate
that TnsA and TnsB interact directly, an interaction that we have long suspected
but which has been difficult to detect. To facilitate structural analysis of the
Tn7 transpososome, we have exploited Tn7-like transposons identified by genome
sequencing. We have found that a TnsAB fusion protein from a cyanobacterium can
carry out transposition in vitro, and we are pursuing the protein's structure.
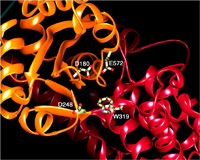
We are also dissecting TnsC, a key regulator of transposition that interacts
with both TnsA and TnsB, as well as the target DNA. Having previously identified
the interacting surfaces of TnsA and TnsB, we are now working to determine how
TnsB and TnsC interact. We have now found that a short segment of TnsC that
interacts with TnsA can activate transposition, a feature we are interested in
exploiting for targeting transposon insertion to particular sites. In contrast to the single transposition protein encoded by hAT and piggyBac
elements, four Tn7-encoded proteins mediate the high-frequency insertion of Tn7
into a specific chromosomal site called attTn7. Tn7 transposition requires the
assembly of an elaborate nucleoprotein complex containing both the donor DNA
from which Tn7 excises and the target DNA into which Tn7 inserts, which contains
all the Tns proteins. Our goal is to understand the structure of this complex
and how its activity is regulated. We now use in vitro assays to demonstrate
that TnsA and TnsB interact directly, an interaction that we have long suspected
but which has been difficult to detect. To facilitate structural analysis of the
Tn7 transpososome, we have exploited Tn7-like transposons identified by genome
sequencing. We have found that a TnsAB fusion protein from a cyanobacterium can
carry out transposition in vitro, and we are pursuing the protein's structure.
We are also dissecting TnsC, a key regulator of transposition that interacts
with both TnsA and TnsB, as well as the target DNA. Having previously identified
the interacting surfaces of TnsA and TnsB, we are now working to determine how
TnsB and TnsC interact. We have now found that a short segment of TnsC that
interacts with TnsA can activate transposition, a feature we are interested in
exploiting for targeting transposon insertion to particular sites.
Cellular Control of and Responses to Transposition
We are also interested in understanding interactions between mobile elements and
their hosts, for example, how the host may regulate transposition, how intact
duplex DNA is generated from the primary products of transposition, and what
impact transposition has upon the host. Since transposition involves the
breakage of DNA molecules, host DNA repair systems are also intimately involved
in transposition. In particular, we are using transposition systems for hAT and
piggyBac elements that we have developed in the highly genetically tractable
organism S. cerevisiae to probe these questions.
 Using Transposons for Genome Analysis and Engineering Using Transposons for Genome Analysis and Engineering
Transposons
are powerful tools for insertional mutagenesis of many organisms and for the
stable introduction of new DNA into a host by transgenesis. We are also
interrogating the organization of the DNA genomes into chromatin by analyzing
high-resolution insertion site profiles of our transposons into the yeast and
human genomes. We have found that both hAT and piggyBac elements insert
preferentially into nucleosome-free regions in yeast, a pattern that we suggest
reflects transposase interaction with the regions of the genome where DNA is
most accessible. These studies identified a heretofore-unknown sequence motif in
nucleosome-depleted regions.
The hyperactive transposons we are isolating and analyzing will contribute to
genome dissection in multiple organisms. For example, we have isolated
hyperactive versions of the mosquito element AeBuster that may facilitate the
now difficult engineering of many insect genomes.
We have also developed a Tn7-based system that mediates site-specific insertion
into bacterial genomes so efficiently that no selection for the insertion event
is required. This is an attractive property for making bacterial strains for
vaccine production. Another significant advance in using transposable elements
for mammalian cell transgenesis would be to develop transposons that will insert
at specific sites so as to avoid unwanted mutagenesis of host genes. We are also
exploring adding sequence-specific DNA-binding domains to hAT and piggyBac
transposases to direct site-specific insertion into mammalian genomes. Also,
there are orthologs of attTn7 in the human genome that Tn7 inserts efficiently
into in vitro, and we are interested in exploiting these sites for element
insertion in vivo.
Domesticated Transposases
 Transposons with a transposase like that of AeBuster1, other hAT elements, and
piggyBac are widely distributed. They also are closely related, however, to
proteins found in the human genome and in some other mammalian genomes. These
mammalian Buster proteins do not appear to be part of transposable elements.
These proteins are extremely similar in sequence, however, strongly suggesting
they have been subject to purifying selection and thus provide important
cellular functions. It is possible that they are "domesticated" transposases
whose ability to bind DNA has been harnessed for some other cellular process—for
example, the regulation of transcription—but which have lost the ability to
promote DNA breakage and joining. We plan to explore the cellular roles of these
proteins. Transposons with a transposase like that of AeBuster1, other hAT elements, and
piggyBac are widely distributed. They also are closely related, however, to
proteins found in the human genome and in some other mammalian genomes. These
mammalian Buster proteins do not appear to be part of transposable elements.
These proteins are extremely similar in sequence, however, strongly suggesting
they have been subject to purifying selection and thus provide important
cellular functions. It is possible that they are "domesticated" transposases
whose ability to bind DNA has been harnessed for some other cellular process—for
example, the regulation of transcription—but which have lost the ability to
promote DNA breakage and joining. We plan to explore the cellular roles of these
proteins. |

