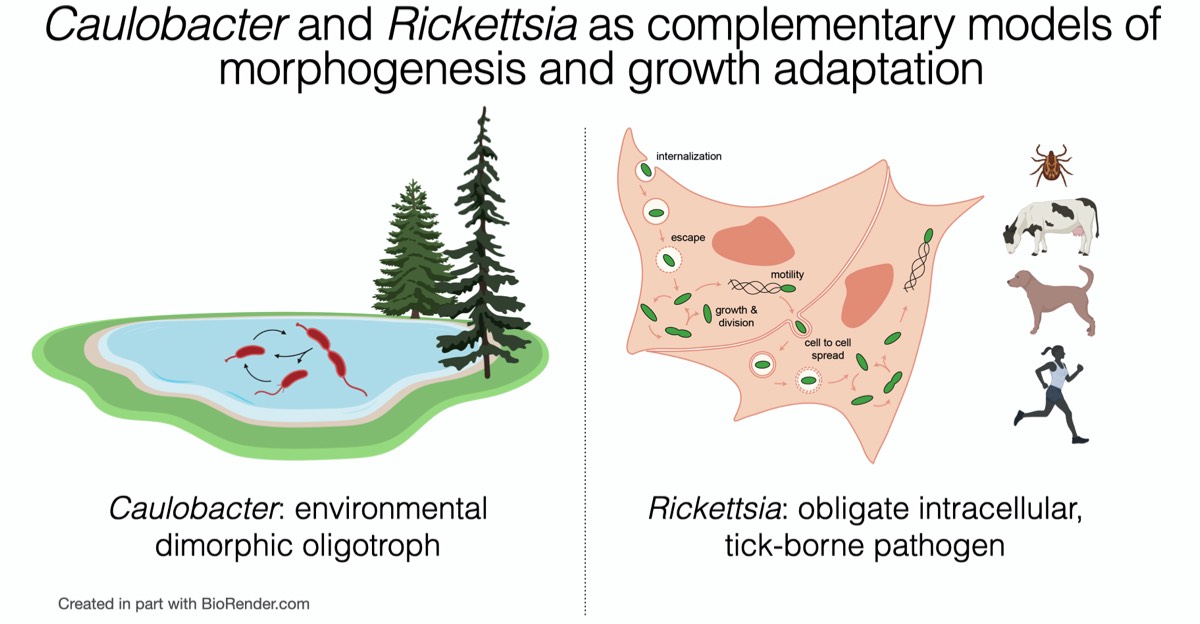
In our laboratory, we take an interdisciplinary approach to study the fundamental mechanisms by which bacteria grow, divide, and adapt to changing growth conditions. We use the dimorphic α-proteobacterium Caulobacter crescentus as one model for these studies, as its curved-rod morphology and well-characterized cell and developmental cycle make it particularly well-suited to address questions of morphogenesis and growth control. We recently initiated studies into a second α-proteobacterium - the obligate intracellular, tick-borne human pathogen Rickettsia parkeri - to investigate the cell biology of the enigmatic Rickettsiales. With their distinct lifestyles and physiology, Caulobacter and Rickettsia present complementary models to understand the diversity of growth and adaptation mechanisms among the α-proteobacteria.
We use a multi-pronged approach combining genetics, genomics, imaging, biochemistry, and in vitro reconstitution to understand the cell biology and adaptation mechanisms of these organisms. We are highly collaborative and love to work with specialists in advanced imaging techniques, new analytical approaches and tools, structural biology, and complementary model organisms. Our fundamental research into essential aspects of bacterial physiology will inform antibiotic development and resistance mechanisms, as well as synthetic cell biology efforts. Currently we are investigating molecular mechanisms by which bacterial growth is regulated at both a global (e.g. during stress) and a local (e.g. at the division site during cytokinesis) level.
We use a multi-pronged approach combining genetics, genomics, imaging, biochemistry, and in vitro reconstitution to understand the cell biology and adaptation mechanisms of these organisms. We are highly collaborative and love to work with specialists in advanced imaging techniques, new analytical approaches and tools, structural biology, and complementary model organisms. Our fundamental research into essential aspects of bacterial physiology will inform antibiotic development and resistance mechanisms, as well as synthetic cell biology efforts. Currently we are investigating molecular mechanisms by which bacterial growth is regulated at both a global (e.g. during stress) and a local (e.g. at the division site during cytokinesis) level.